Bio Endeavors, August 2022: Harnessing evolution
The ingenuity of evolution
Life has found a way to, well, live. While scientists and philosophers alike have long debated what it means to be alive, the idea that life is a self-sustained system capable of undergoing Darwinian evolution is remarkably powerful. Simply put, to be alive is to be part of an engine that selects traits with survival advantages over many, many generations. Over time, living systems have become extraordinarily complex. In our last post, we highlighted this complexity in the context of plant biology and the chemical diversity that evolution has naturally generated1.
Finding solutions to major engineering biology problems today — developing new medicines, growing foods in novel ways, developing climate-resilient crops — is really challenging. Engineers design solutions through a rigorous optimization process. In the context of biology, however, designing is not always possible because we don’t know all the rules. As a result, we mostly discover. But, as we’ve seen over the last 3.7 billion years, evolutionary processes are uniquely powerful tools. In the context of building biological systems, what can we learn and utilize?
One of the earliest cited examples of co-opting functionality from microbes (excluding ancient practices for alcohol and cheese) is the discovery of a fungus during World War II. While stationed on the Solomon Islands in the South Pacific, the US Army was baffled by the troublesome deterioration of their tents, clothing, and other textiles. After taking samples and then screening over 14,000 molds recovered from the site, researchers at Natick Army Research Laboratories found the culprit: a strain of fungus, now known as Trichoderma reesei, which produced extracellular enzymes (i.e., cellulases) that degraded their textiles. This turned out to be a massively important discovery in the quest to convert biomass to biofuels: the enzymes that T. reesei produced degraded not only textiles and recalcitrant biomass (lignocellulose), in turn releasing sugars that could be further converted into fuel. However, performance was still not good enough for industrial production, so in the 1970s, researchers began work to further increase the efficiency of T. reesei. What followed were some of the original directed evolution methods whereby researchers introduced random mutagenesis by blasting the bugs with radiation and then screened the mutants in a functional assay. Using these experiments, they were able to ~3x the performance of the bug, creating the foundation for the gold standard strains we have today.
Today, directed evolution experiments are ingrained in the scientific canon: notably, Frances Arnold shared the Nobel Prize in 2018 for her ingenious approach of directed evolution to engineer enzymes. And yet, history has only scratched the surface of what directed evolution experiments can do. New tools are beginning to revolutionize what is possible. For example:
New “writing” tools are expanding the types of problems we can tackle: Increasingly, we can be more targeted with how we edit enzymes, microbes, or populations of microbes to generate diversity. Or we can engineer entirely novel genetic circuits that enable new objective functions, for example, making a bug unable to survive unless it produces a specific compound.
New “reading” tools help us select winners: With better sensing technologies (e.g., biosensors, spectrometry, single cell multi-omics), we can identify and select across a greater range of phenotypes with higher throughput. In the case of strain engineering in industrial biomanufacturing, this is particularly important for selecting a phenotype other than simple growth.
New “optimization” tools help us drive continuous improvement: With novel bioreactors and automation, we can run a much higher number of evolutionary experiments in parallel and precisely adjust environmental conditions for each to drive improved performance over time. Additionally, improved computational approaches can help us learn from these experiments and optimize across many variables simultaneously. Iteratively running these experiments can help us guide evolution through a multidimensional design space with unprecedented precision.
So what? If early directed evolution experiments were like a simple iterative algorithm, tomorrow’s directed evolution experiments can be much more like a black-box ML approach. We are quickly moving towards a world where we can program complex objective functions into biological systems, and build methods that let those systems solve the problems for us. Today, we are exploring this topic of directed evolution and how we might use new methods to solve problems across two applications: industrial synthetic biology and drug discovery.
Industrial synthetic biology: Engineering for stability and scale
Historical context: In industrial synthetic biology, we design microbes as factories for producing molecules of interest: everything from heme in Impossible Foods’ burgers to proteases for fabric detergent to cellulases as in Natick Army Research Laboratories. Microbes make great factories because they self-replicate, scale relatively quickly, and can be tuned to replace many existing, dirty manufacturing processes. The idea is straightforward: we should just program bugs to produce whatever we need in large industrial fermenters.
Companies have been working on this (seemingly simple) problem for decades. Genentech started the recombinant protein trend in the late 70s with somatostatin followed by insulin, and the rigorous engineering discipline picked up a lot of momentum at the turn of the century with the emergence of genomics and systems biology. More recently, companies like Cargill, IFF, Amyris, Ginkgo, and Zymergen2 play important roles in the ecosystem. Nevertheless, economic targets, especially in existing commodity markets, demand massive scale and leave little margin for error. Unfortunately, evolution poses a big problem in industrial biology. Microbial populations move quickly to do what life does best: rapidly evolve for survival, often in unpredictable ways that are highly sensitive to changing conditions (e.g., a scale-up from benchtop to >100k L bioreactors) and to the detriment of the economic targets (e.g., titer, doubling time) we set.
Unfortunately, bottom-up rational design tools (i.e., leveraging known parts and traits) 3have been unable to manage the complex tradeoffs between fitness under varying conditions and the production of the things we care about. Andrew Horwitz of Sestina Bio has a great blog post that discusses this tradeoff in greater detail.
Optimizing with evolution in mind (a brief review of helpful literature): Excitingly, new approaches are emerging that steer evolution in ways that better account for these tradeoffs (at both individual and population levels). To frame the environment and design space we’re operating in when we engineer living systems, Castle et al. introduced several concepts.
Most saliently, they describe the evotype - the evolutionary disposition of the system — which is a critical design component. Per the figure shown above, the evotype is a function of the “fitneity” of the system (i.e., reproductive fitness x desired utility), the phenotype (measured by desired utility), and the variation probability distribution (i.e., how likely is this “fitneity” phenotype). This landscape needs to be designed and traversed in biological systems design to ensure a stable and desired phenotype is reached. In a more tactical review, Sánchez et al. provides an overview of how tools of directed evolution might be applied to engineer microbial ecosystems4. Most of our directed evolution experiments today work in the context of a single bug or enzyme, but engineering ecosystems is incredibly relevant for industrial synthetic biology where we are working on the scale of ecosystems and need robust population dynamics. Similar to the conceptual framework described in Castle et al., this work imagines how to traverse the ecological landscapes to arrive at robust, stable, and desired phenotypes. Importantly, they describe the characteristics required for engineering these systems as:
Phenotypic variations are distributed along a vector of selection that is heritable.
Mechanisms that introduce variability and explore structure-function landscapes.
A community size that enables stochastic sampling for between-population variability.
Enough experimental time to reach a stable, fixed population.
The benefit of using evolutionary processes to solve these problems is that they can deliver traits to bugs that are fitness maxima and therefore stabilized and maintained by selection. This is in contrast to designed traits, which are often eroded by selection. This is an important characteristic of these populations because of the requirements to scale up production. Ultimately, the larger the fermenter, the more generations of selection, and the more potential for the populations of strains to veer off course. Processes that design for evolutionary dynamics can be more robust to the pressures of scale.
What we’re excited to see: Building on the existing infrastructure described in the reviews previously mentioned, we envision new tools and infrastructure will be critical for bringing these approaches to bear at scale in the world. We anticipate an integrated evolutionary design system will require a few elements:
Further development of hardware and wetware to drive throughput and control: An automated system that is capable of running many experiments in parallel would be helpful. This would need to be capable of running long experiments that reflect the scaled industrial setting and enable effective exploration of the design space. These reactors would be just large enough to support relevant population sizes, have independent, controlled environments, and be measured distinctly with minimally (or non-destructive) approaches.
Novel measurement tools to generate necessary data: The specific measurement tools are an area we need further investment. Today, measuring many phenotypes beyond growth without destructive systems is hard and expensive. High-content imaging approaches (like Raman) are exciting in this context.
Novel computational approaches to orchestrate the system and navigate the design space, ultimately driving nonlinear improvement and scale over time.
In a startup context, thoughtful integration with other tools in the strain development stack and paths to capture meaningful value (more on this in a future newsletter).
Drug discovery: Programming evolution to solve for function
Historical context: We use the term “drug discovery” rather than “drug design” for understandable reasons. Historically biomedical researchers have discovered small molecule drugs through a serendipitous game of brute force. Once we identify a target, we usually throw a bunch (read: millions) of molecules at it to see what sticks. This whole process is referred to as high-throughput screening. We have had the most success working on the canonical drug targets with well-defined and distinct small molecule binding pockets in their active sites. These canonical targets are sometimes referred to as the druggable proteome, though specific definitions of druggable aren’t specific and are constantly changing (e.g., as in kinases).
On the more in silico side, we’ve seen great work building generative chemistry approaches from investigators like Connor Coley, Regina Barzilay, and Gisbert Schneider, that have built hybrid computational-experimental methods. These show strong promise in generating novel chemical matter more efficiently than traditional approaches. That said, bottoms-up, rational design approaches are still early in generating results that translate to the clinic, especially in challenging, undruggable targets. There are a variety of reasons for this:
Native context matters and remains complex to accurately model:
Biology is so context-dependent in how it is expressed, and therefore, not served well by reductionist approaches. Despite the emergence of concepts like digital biology as an analogy for the principles of synthetic biology, it is important to note that biology is distinctly non-digital5. Cells and organisms are incredibly efficient machines, but their constituent parts are not. The emergent properties of these systems are highly sophisticated, but the core components are sloppy and error-prone. So, studying the components in isolation, as we mostly do in high-throughput screening in biochemical assays, makes the data easier to generate, but limits translatability to the clinic.
Ideally, you’d be able to isolate the target you want to study but have the ability to view the effect in a native cellular context. Eikon Therapeutics, for example, directly visualizes protein kinetics in cell-based models and can utilize this approach in high-throughput screening assays (read more in our blog posts here and here).
Structure-based drug design assumes a well-defined and modellable structure:
AlphaFold by Deepmind has made an enormous leap forward in our ability to predict protein structure from its amino acid sequence alone (if you really want to dive down a thrilling science rabbit hole, it’s worth digging into the post by Deepmind and following all the linked resources). This achievement has opened up tons of amazing downstream applications, but it hasn’t solved drug discovery. Cue Derek Lowe with a more sobering take on the AlphaFold achievement and what it means for drug discovery.
A large part of this is because AlphaFold and other structure-based approaches cannot capture regions of proteins that are unstructured (estimated at 37-50% of proteome) and certainly struggle with targets that don’t have well-defined binding pockets. Many undruggable targets fall in this category; these include transcription factors, non-enzymatic proteins, protein-protein interactions, and phosphatases, among others. Estimates place the percentage of undruggable targets at ~85% (though this number is constantly changing; kinases were once considered undruggable and now are a well-trodden class of targets).
Chemical diversity is really important, but it’s hard to find and presents a synthesis challenge:
Today’s high-throughput screening libraries are biased toward historical successes (as are the in silico models built on these training data). There is a reinforcement loop where success begets more success, and these libraries aren’t super diverse as a result, especially so for undruggable targets without historical precedence. Additionally, synthetic accessibility is always challenging with generative small molecule drug design. Natural products are a great option for chemical diversity (as we’ve alluded to with plant chemistry) but are often challenging to source or synthesize. Artemisinin, the anti-malarial drug, is a famous example of the challenges of scaling up synthesis.
Evolution-driven approaches as an alternative: As in the prior example, where our understanding of biology falls short, evolution provides a potential solution. Evolution-driven approaches to solve these problems are in some ways analogous to a biological black-box ML algorithm: set an objective function for the microbe, for example, inhibition of a target, parameterize the search space, and let the microbe evolve to solve for a given function in ways that are often novel or not intuitive.
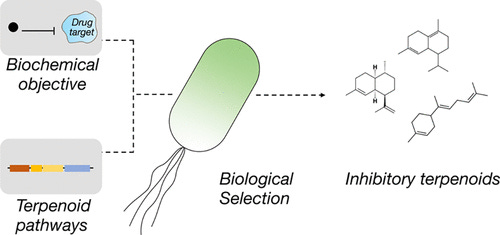
In an interesting example, Sarkar et al. built a system that does just this. They programmed an objective function into a microbe by requiring that the microbe inhibit an undruggable target, protein tyrosine phosphatase 1B (PTP1B), to survive6. They also gave the bug the tools for the job: in this case, adding pathways to enable the biosynthesis of terpenoids, a large class of natural products7.
The project was a great proof of concept of this approach. The investigators:
Identified two previously unknown terpenoid inhibitors of the PTP1B (the drug target).
The investigators then found a novel binding mode that differs from a previously characterized allosteric inhibitor (and thus not one we would have known to design for); this mode was further characterized in subsequent work, which showed engagement by a disordered target region.
Lastly, they demonstrated the value of leveraging the microbe as the synthesis engine for the compounds; this can be a powerful approach for utilizing the chemical diversity attributed to natural products.
What we’re excited to see: Leveraging evolutionary processes in these ways represents an exciting future direction of drug discovery, combining trends of natural product chemistry, functional assays, allostery, and undruggable targets — equipping bugs with the tools of plants to augment medicinal chemists. We’re a long way off from fully automating medicinal chemistry, but bugs and biosynthesis may represent an interesting way to get there.
What we’re reading & listening to
Looking into evolution
💻Using generative models of evolution to predict disease variants and design proteins → How can evolution inform novel computational approaches?
🧬Building an RNA switch-based selections system for enzyme evolution in Yeast → In directed evolution processes, selection for diverse phenotypes is challenging; RNA switches represent a modular frontier.
🧬 Biosynthesis of saponin defensive compounds in sea cucumbers → What’s slower than a sea cucumber? Mostly nothing, which is why they need interesting chemistry to defend themselves.
🧬 Low-cost anti-mycobacterial drug discovery using engineered E. coli → Similar to the example described previously, leveraging microbes as a functional assay to identify novel Mycobacterium tuberculosis inhibitors.
Also on our minds
🧬Tech-driven synthetic biology: building a cloud-first modern wet lab → Great example of tightly coordinating wet-lab and in silico work, featuring Peyton and Ed at BigHat Biosciences.
No chemical killer AI (yet) → A helpful reminder of the importance of biosafety
💻Controllable protein design with language models → Helpful review on how NLP research is influencing protein science.
🧬Engineering consortia by polymeric microbial swarmbots → Microbial consortia display super interesting emergent properties.
Genome-wide CRISPR screens of T cell exhaustion identify chromatin remodeling factors that limit T cell persistence → A step forward for a large problem in cancer immunotherapy.
🌎Biodiversity mediates ecosystem sensitivity to climate change → Perhaps biodiversity and climate are more interdependent than we thought.
👽First images from the James Webb Space Telescope (and great podcast about it)→ Not biology but mind-blowing.
Want more from Innovation Endeavors?
Subscribe to our blog today for more updates and insights from our portfolio and our team!
Questions? Comments? Ideas?
We are always excited to engage with scientists, educators, investors, industry veterans, and entrepreneurs tackling essential challenges with biology. If you have feedback, questions, thoughts, or ideas, drop us a note at bio@innovationendeavors.com.
Special thank you to Sam Levin (Melonfrost), Gaurav Venkataraman (Trisk Bio), Jerome Fox and Ankur Sarkar (Think Bio), Jess McKinley (Plenty), Darren Platt (Demetrix), Alex Patist (Geltor) for conversations that have contributed to this newsletter, and to all the scientists whose work we have referenced. We appreciate the creative solutions you are bringing to these important problems. Let’s continue to evolve.
We look forward to hearing from you. Until next time!
- The Innovation Endeavors bio team: Dror, Sam, Nick, and Carrie
Plants are such an interesting segment of life because they’re immobile and therefore unable to physically avoid predators and scavenge for food. This necessity has created massive amounts of chemical diversity — plants, together, produce over a million different small molecule chemicals that they use to grow, communicate, and protect themselves from predators.
Recently acquired by Gingko.
We’re drawing a distinction between what Sánchez et al. describes as top-down vs bottom-up engineering. Bottom-up refers to working with already known parts and traits; top-down does not require any a priori knowledge of the mechanisms responsible for the desired functionality.
Several of the authors push this even further in separate work by building a flexible modeling framework that demonstrates the feasibility of directed evolution approaches to drive top-down engineering of microbial consortia; separate but related frameworks by Xie and Shou and Lalejini et al. are also worth reviewing.
That is not to say there is not a lot to learn from traditional computing in the way of standard parts, abstractions, etc., but to do it well, we must respect the substrate. In many ways, this newsletter is highlighting the unique aspects of the substrate in solving our challenges.
Phosphatases, as previously alluded to, squarely fall in this camp of the undruggable. No approved therapeutics target a phosphatase; that said, it is a class of much interest. SH2 containing protein tyrosine phosphatase-2 (SHP2) is a phosphatase that many are going after; early data suggests that SHP2 plays a role in tumor microenvironment and sensitivity to I/O therapy.
Natural product pathways — terpenoids, alkaloids, phenols, etc. — provide an amazing diversity of chemistry, and have been leveraged for a long time in the form of ancient, natural medicines and modern pharma drug development; more famous examples include paclitaxel, artemisinin, digitalis.